Potential evapotranspiration under warming and elevated \(CO_2\) concentration#
M. Vremec, R.A. Collenteur University of Graz, 2021
In this notebook it is shown how to estimate Potential Evapotranspiration under elevated \(CO_2\) conditions and an increase in temperature. The work follows the suggestions made by Yang et al. (2019) to include the sensitivity of stomatal resistance to \(CO_2\) in the Penman-Monteith equation.
Yang, Y., Roderick, M.L., Zhang, S. et al. Hydrologic implications of vegetation response to elevated CO2 in climate projections. Nature Clim Change 9, 44–48 (2019). https://doi.org/10.1038/s41558-018-0361-0
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import pyet as pyet
1. Load daily data from ZAMG (Messstationen Tagesdaten)#
station: Graz Universität 16412
Selected variables:
globalstrahlung (global radiation), J/cm2 needs to be in MJ/m3d, ZAMG abbreviation - strahl
arithmetische windgeschwindigkeit (wind speed), m/s, ZAMG abbreviation - vv
relative feuchte (relative humidity), %, ZAMG abbreviation - rel
lufttemparatur (air temperature) in 2 m, C, ZAMG abbreviation - t
lufttemperatur (air temperature) max in 2 m, C, ZAMG abbreviation - tmax
lufttemperatur (air temperature) min in 2 m, C, ZAMG abbreviation - tmin
latitute and elevation of a station
#read data
data_16412 = pd.read_csv('data/example_1/klima_daily.csv', index_col=1, parse_dates=True)
data_16412
| station | strahl | rel | t | tmax | tmin | vv | |
|---|---|---|---|---|---|---|---|
| time | |||||||
| 2000-01-01 | 16412 | 300.0 | 80.0 | -2.7 | 0.5 | -5.8 | 1.0 |
| 2000-01-02 | 16412 | 250.0 | 86.0 | 0.2 | 2.5 | -2.1 | 1.0 |
| 2000-01-03 | 16412 | 598.0 | 86.0 | 0.6 | 3.6 | -2.4 | 1.0 |
| 2000-01-04 | 16412 | 619.0 | 83.0 | -0.5 | 4.5 | -5.5 | 1.0 |
| 2000-01-05 | 16412 | 463.0 | 84.0 | -0.1 | 5.4 | -5.5 | 1.0 |
| ... | ... | ... | ... | ... | ... | ... | ... |
| 2021-11-07 | 16412 | 852.0 | 74.0 | 8.5 | 12.2 | 4.7 | 1.6 |
| 2021-11-08 | 16412 | 553.0 | 78.0 | 7.5 | 10.4 | 4.5 | 1.6 |
| 2021-11-09 | 16412 | 902.0 | 67.0 | 7.1 | 11.7 | 2.4 | 2.7 |
| 2021-11-10 | 16412 | 785.0 | 79.0 | 5.3 | 10.1 | 0.4 | 2.1 |
| 2021-11-11 | 16412 | 194.0 | 91.0 | 5.1 | 6.5 | 3.7 | 1.1 |
7986 rows × 7 columns
# Convert Glabalstrahlung J/cm2 to MJ/m2 by dividing to 100
meteo = pd.DataFrame({"time":data_16412.index, "tmean":data_16412.t, "tmax":data_16412.tmax, "tmin":data_16412.tmin, "rh":data_16412.rel,
"wind":data_16412.vv, "rs":data_16412.strahl/100})
time, tmean, tmax, tmin, rh, wind, rs = [meteo[col] for col in meteo.columns]
lat = 47.077778 * np.pi / 180 # Latitude of the meteorological station, converting from degrees to radians
elevation = 367 # meters above sea-level
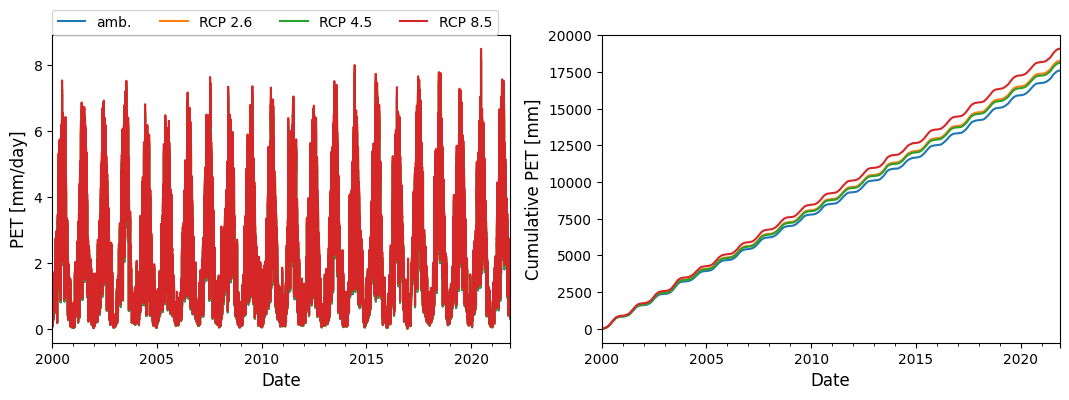
2.Estimate potential evapotranspiration under RCP scenarios#
# the first element in the list includes the rise in temperature, and the CO2 concentration
rcp_26 = [1.3, 450]
rcp_45 = [2, 650]
rcp_85 = [4.5, 850]
def pe_cc(tincrease, co2):
pe = pyet.pm(tmean+tincrease, wind, rs=rs, elevation=elevation, lat=lat,
tmax=tmax+tincrease, tmin=tmin+tincrease, rh=rh, co2=co2)
return pe
# Estimate evapotranspiration with four different methods and create a dataframe
pet_df = pd.DataFrame()
pet_df["amb."] = pe_cc(0, 420)
pet_df["RCP 2.6"] = pe_cc(rcp_26[0], rcp_26[1])
pet_df["RCP 4.5"] = pe_cc(rcp_45[0], rcp_45[1])
pet_df["RCP 8.5"] = pe_cc(rcp_85[0], rcp_85[1])
fig, axs = plt.subplots(figsize=(13,4), ncols=2)
pet_df.plot(ax=axs[0])
pet_df.cumsum().plot(ax=axs[1], legend=False)
axs[0].set_ylabel("PET [mm/day]", fontsize=12)
axs[1].set_ylabel("Cumulative PET [mm]", fontsize=12)
axs[0].legend(ncol=6, loc=[0,1.])
for i in (0,1):
axs[i].set_xlabel("Date", fontsize=12)